At this stage, basic requirements such as Java 1.6+ JDK, Hadoop and Oozie installations should be available. The following brief documentation will explain working with Oozie workflows.
Copy the workflow application directory to your HDFS. ($HADOOP_HOME/bin should be in command path)
$ hadoop fs -put <src path on local file system> <destination path>
A workflow application directory has the following structure
A coordinator application directory has a 'coordinator.xml' file in addition to the above.
Oozie workflow enables you to execute your task via multiple action options e.g. Java action, Map-Reduce action, Pig action, Fs action and so on.
Oozie jobs are executed on the Hadoop cluster via a Launcher (Refer to section Launcher-Mapper on this page). Hence the workflow has to be configured with the parameters-
For example, the usage without Oozie for submitting a hadoop job on CLI is,
$ hadoop [COMMAND] [GENERIC_OPTIONS]
The GENERIC_OPTIONS comprise
Now with Oozie, the equivalent properties can be specified
<job-xml> ... </job-xml>
<name-node> ... </name-node>
<job-tracker> ... </job-tracker>
<files> ... </files>
<archives> ... </archives>
<configuration>
<property>
<name> ... </name>
<value> ... </value>
</property>
</configuration>
OR
Note: The job.properties files need not be uploaded to HDFS as part of the workflow app directory. It is only required locally on the machine from where oozie job is submitted. That way you can use various property values to submit same job to different cluster environments.
Sample job.properties file
nameNode=foo:9000
jobTracker=bar:9001
jobInput=/somedirpath
queueName=default
Sample Syntax for the workflow.xml file with a Java action (illustrating use of EL expressions from job.properties).
<workflow-app name="[WF-DEF-NAME]" xmlns="uri:oozie:workflow:0.2">
...
<action name="[NODE-NAME]">
<java>
<job-tracker>${jobTracker}</job-tracker>
<name-node>${nameNode}</name-node>
<prepare>
<delete path="[PATH]"/>
...
<mkdir path="[PATH]"/>
...
</prepare>
<configuration>
<property>
<name>mapred.job.queue.name</name>
<value>${queueName}</value>
</property>
<property>
<name>mapred.input.dir</name>
<value>${jobInput}</value>
</property>
...
</configuration>
<main-class>[MAIN-CLASS]</main-class>
<java-opts>[JAVA-STARTUP-OPTS]</java-opts>
<arg>ARGUMENT</arg>
...
</java>
<ok to="[NODE-NAME]"/>
<error to="[NODE-NAME]"/>
</action>
...
</workflow-app>
The syntax of the tags remains the same for Java, Map-Reduce, Pig, Fs or Ssh actions in Oozie.
There are different ways of parameterization of configuration values, by passing them in workflow.xml, job.properties, config-default.xml, or a custom xml file referred to via the job-xml tag in the workflow. For more details refer to the section How to Parameterize Oozie Jobs.
A workflow action can be configured to perform HDFS files/directories cleanup before starting the application. This capability enables Oozie to retry an application in the situation of a transient or non-transient failure (This can be used to cleanup any temporary data which may have been created by the application in case of failure).
The prepare element, if present, indicates a list of paths to do file operations upon, before starting the application. This should be used exclusively for directory cleanup for the application to be executed; only delete and mkdir operations can be done in order.
<prepare>
<delete path=[PATH] />
..
<mkdir path=[PATH] />
..
</prepare>
It is possible to add files and archives as workflow elements to be available to the application. If the specified path is relative, it is assumed the file or archive are within the application directory, in the corresponding sub-path. If the path is absolute, the file or archive it is expected in the given absolute path. These files are copied to the map reduce cluster compute node. The archives specified in the arguments list are unarchived on the compute machines.
Files specified with the file element, will be symbolic links in the current working directory i.e. home directory of the task. If a file is a native library (an '.so' or a '.so.#' file), it will be symlinked as an '.so' file in the task running directory, thus available to the task JVM. To force a symlink for a file on the task running directory, use a '#' followed by the symlink name. (Illustrated below)
Oozie supports these by allowing file and archive tags that can be defined in the application workflow as below:
<file> dir1/dict.txt#dict1 </file>
<file> dir2/dict.txt#dict2 </file>
<archive> mytar.tgz#tgzdir </archive>
Here, the files dir1/dict.txt and dir2/dict.txt can be accessed by jobs using the symbolic names dict1 and dict2 respectively. The archive mytar.tgz will be placed and unarchived into a directory by the name "tgzdir".
Please note that the addlibjars option supported by the Hadoop command-line is not supported by Oozie.
A common misunderstanding among the users is that the Oozie-server launches the MapReduce/Pig jobs by itself. The following diagram shows what actually happens when Oozie tries to launch actions in a workflow:
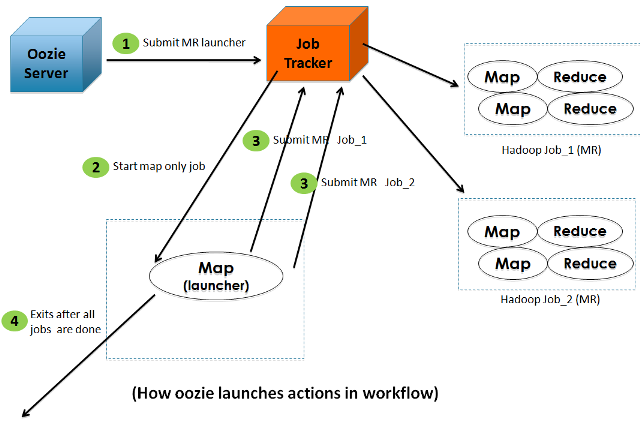
The reasons for using a Launcher job as an intermediate step are:
Now to begin translating your tasks into equivalent Oozie jobs by utilizing different actions and writing workflows incorporating the above features, refer to Workflow Specifications comprising of different Oozie Actions.
Detailed use-cases and composition :-