- General
- Developers
- Mahout-Samsara
- Algorithms
- List of algorithms
- Distributed Matrix Decomposition
- Cholesky QR
- SSVD
- Distributed ALS
- SPCA
- Recommendations
- Recommender Overview
- Intro to cooccurrence-based
recommendations with Spark - Classification
- Spark Naive Bayes
- MapReduce Basics
- List of algorithms
- Overview
- Working with text
- Creating vectors from text
- Collocations
- Dimensionality reduction
- Singular Value Decomposition
- Stochastic SVD
- Topic Models
- Latent Dirichlet Allocation
- Mahout MapReduce
- Classification
- Naive Bayes
- Hidden Markov Models
- Logistic Regression (Single Machine)
- Random Forest
- Classification Examples
- Breiman example
- 20 newsgroups example
- SGD classifier bank marketing
- Wikipedia XML parser and classifier
- Clustering
- k-Means
- Canopy
- Fuzzy k-Means
- Streaming KMeans
- Spectral Clustering
- Clustering Commandline usage
- Options for k-Means
- Options for Canopy
- Options for Fuzzy k-Means
- Clustering Examples
- Synthetic data
- Cluster Post processing
- Cluster Dumper tool
- Cluster visualisation
- Recommendations
- First Timer FAQ
- A user-based recommender
in 5 minutes - Matrix factorization-based
recommenders - Overview
- Intro to item-based recommendations
with Hadoop - Intro to ALS recommendations
with Hadoop
k-Means clustering - basics
k-Means is a simple but well-known algorithm for grouping objects, clustering. All objects need to be represented as a set of numerical features. In addition, the user has to specify the number of groups (referred to as k) she wishes to identify.
Each object can be thought of as being represented by some feature vector in an n dimensional space, n being the number of all features used to describe the objects to cluster. The algorithm then randomly chooses k points in that vector space, these point serve as the initial centers of the clusters. Afterwards all objects are each assigned to the center they are closest to. Usually the distance measure is chosen by the user and determined by the learning task.
After that, for each cluster a new center is computed by averaging the feature vectors of all objects assigned to it. The process of assigning objects and recomputing centers is repeated until the process converges. The algorithm can be proven to converge after a finite number of iterations.
Several tweaks concerning distance measure, initial center choice and computation of new average centers have been explored, as well as the estimation of the number of clusters k. Yet the main principle always remains the same.
Quickstart
Here is a short shell script outline that will get you started quickly with k-means. This does the following:
- Accepts clustering type: kmeans, fuzzykmeans, lda, or streamingkmeans
- Gets the Reuters dataset
- Runs org.apache.lucene.benchmark.utils.ExtractReuters to generate reuters-out from reuters-sgm (the downloaded archive)
- Runs seqdirectory to convert reuters-out to SequenceFile format
- Runs seq2sparse to convert SequenceFiles to sparse vector format
- Runs k-means with 20 clusters
- Runs clusterdump to show results
After following through the output that scrolls past, reading the code will offer you a better understanding.
Implementation
The implementation accepts two input directories: one for the data points and one for the initial clusters. The data directory contains multiple input files of SequenceFile(Key, VectorWritable), while the clusters directory contains one or more SequenceFiles(Text, Cluster) containing k initial clusters or canopies. None of the input directories are modified by the implementation, allowing experimentation with initial clustering and convergence values.
Canopy clustering can be used to compute the initial clusters for k-KMeans:
// run the CanopyDriver job
CanopyDriver.runJob("testdata", "output"
ManhattanDistanceMeasure.class.getName(), (float) 3.1, (float) 2.1, false);
// now run the KMeansDriver job
KMeansDriver.runJob("testdata", "output/clusters-0", "output",
EuclideanDistanceMeasure.class.getName(), "0.001", "10", true);
In the above example, the input data points are stored in ‘testdata’ and the CanopyDriver is configured to output to the ‘output/clusters-0’ directory. Once the driver executes it will contain the canopy definition files. Upon running the KMeansDriver the output directory will have two or more new directories: ‘clusters-N’’ containining the clusters for each iteration and ‘clusteredPoints’ will contain the clustered data points.
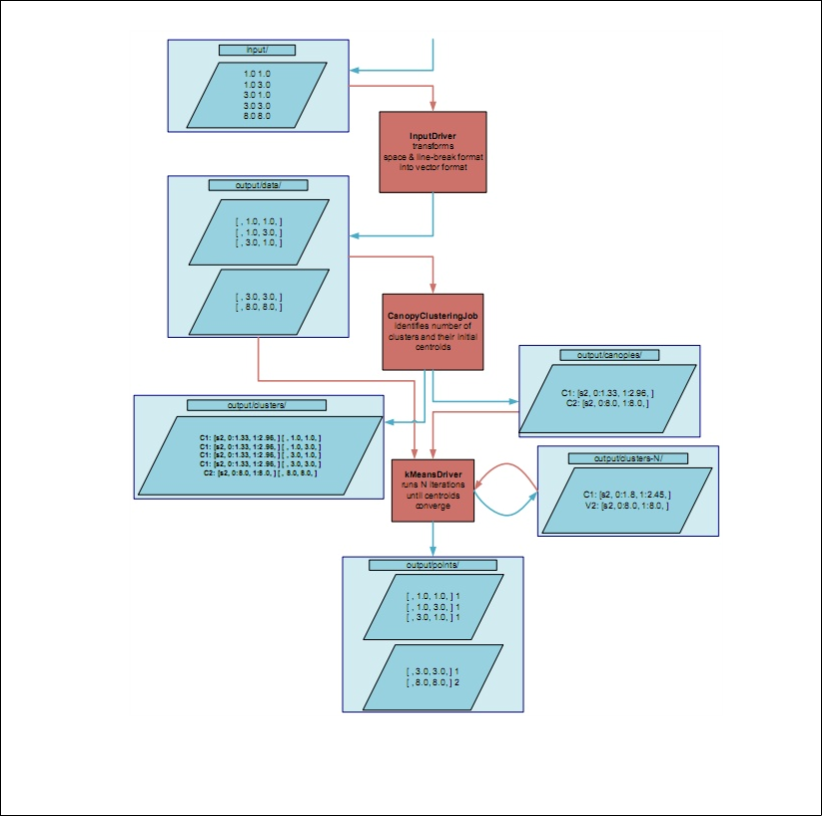
This diagram shows the examplary dataflow of the k-Means example
implementation provided by Mahout:
Running k-Means Clustering
The k-Means clustering algorithm may be run using a command-line invocation on KMeansDriver.main or by making a Java call to KMeansDriver.runJob().
Invocation using the command line takes the form:
bin/mahout kmeans \
-i <input vectors directory> \
-c <input clusters directory> \
-o <output working directory> \
-k <optional number of initial clusters to sample from input vectors> \
-dm <DistanceMeasure> \
-x <maximum number of iterations> \
-cd <optional convergence delta. Default is 0.5> \
-ow <overwrite output directory if present>
-cl <run input vector clustering after computing Canopies>
-xm <execution method: sequential or mapreduce>
Note: if the -k argument is supplied, any clusters in the -c directory will be overwritten and -k random points will be sampled from the input vectors to become the initial cluster centers.
Invocation using Java involves supplying the following arguments:
- input: a file path string to a directory containing the input data set a SequenceFile(WritableComparable, VectorWritable). The sequence file key is not used.
- clusters: a file path string to a directory containing the initial clusters, a SequenceFile(key, Cluster | Canopy). Both KMeans clusters and Canopy canopies may be used for the initial clusters.
- output: a file path string to an empty directory which is used for all output from the algorithm.
- distanceMeasure: the fully-qualified class name of an instance of DistanceMeasure which will be used for the clustering.
- convergenceDelta: a double value used to determine if the algorithm has converged (clusters have not moved more than the value in the last iteration)
- maxIter: the maximum number of iterations to run, independent of the convergence specified
- runClustering: a boolean indicating, if true, that the clustering step is to be executed after clusters have been determined.
- runSequential: a boolean indicating, if true, that the k-means sequential implementation is to be used to process the input data.
After running the algorithm, the output directory will contain:
- clusters-N: directories containing SequenceFiles(Text, Cluster) produced by the algorithm for each iteration. The Text key is a cluster identifier string.
- clusteredPoints: (if --clustering enabled) a directory containing SequenceFile(IntWritable, WeightedVectorWritable). The IntWritable key is the clusterId. The WeightedVectorWritable value is a bean containing a double weight and a VectorWritable vector where the weight indicates the probability that the vector is a member of the cluster. For k-Means clustering, the weights are computed as 1/(1+distance) where the distance is between the cluster center and the vector using the chosen DistanceMeasure.
Examples
The following images illustrate k-Means clustering applied to a set of randomly-generated 2-d data points. The points are generated using a normal distribution centered at a mean location and with a constant standard deviation. See the README file in the /examples/src/main/java/org/apache/mahout/clustering/display/README.txt for details on running similar examples.
The points are generated as follows:
- 500 samples m=[1.0, 1.0](1.0,-1.0.html) sd=3.0
- 300 samples m=[1.0, 0.0](1.0,-0.0.html) sd=0.5
- 300 samples m=[0.0, 2.0](0.0,-2.0.html) sd=0.1
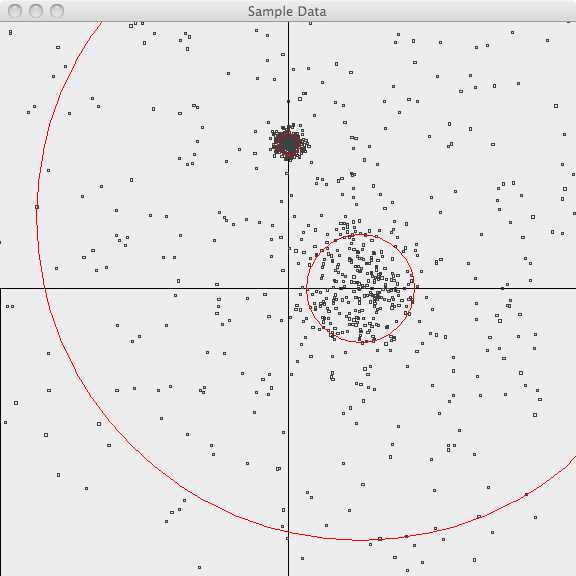
In the first image, the points are plotted and the 3-sigma boundaries of their generator are superimposed.
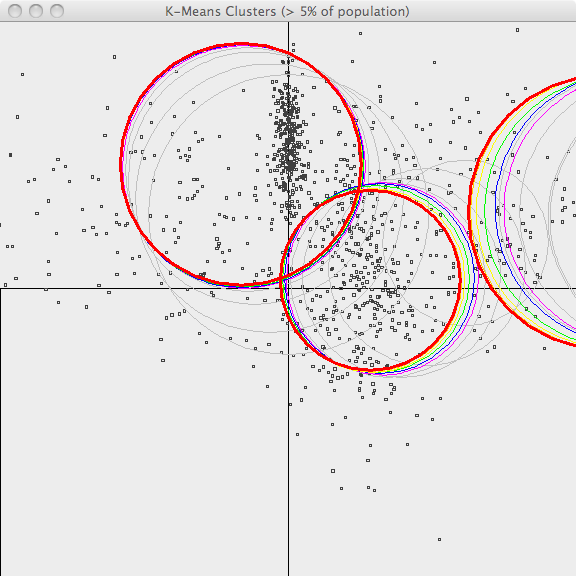
In the second image, the resulting clusters (k=3) are shown superimposed upon the sample data. As k-Means is an iterative algorithm, the centers of the clusters in each recent iteration are shown using different colors. Bold red is the final clustering and previous iterations are shown in [orange, yellow, green, blue, violet and gray](orange,-yellow,-green,-blue,-violet-and-gray.html) . Although it misses a lot of the points and cannot capture the original, superimposed cluster centers, it does a decent job of clustering this data.
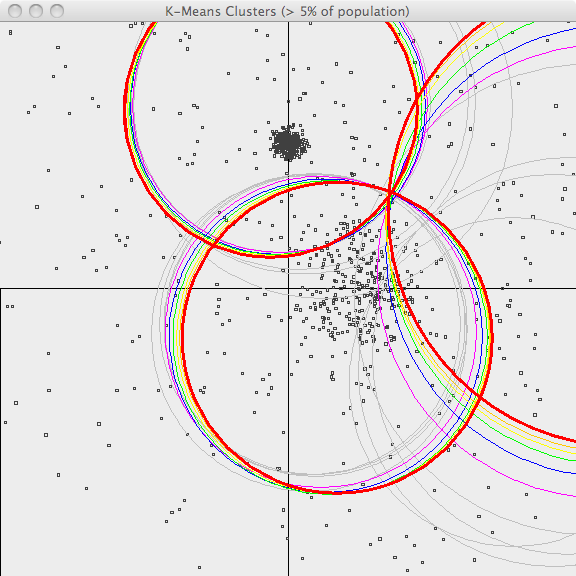
The third image shows the results of running k-Means on a different dataset, which is generated using asymmetrical standard deviations. K-Means does a fair job handling this data set as well.